10.13 Solutions
Solution to Exercise 10.12.1:
## Call: survfit(formula = survival_object ~ ulcer, data = melanoma)
##
## ulcer=No
## time n.risk n.event survival std.err lower 95% CI upper 95% CI
## 0 115 0 1.000 0.0000 1.000 1.000
## 1 112 2 0.983 0.0122 0.959 1.000
## 2 112 0 0.983 0.0122 0.959 1.000
## 3 107 5 0.939 0.0225 0.896 0.984
## 4 105 2 0.921 0.0252 0.873 0.972
## 5 78 4 0.883 0.0306 0.825 0.945
##
## ulcer=Yes
## time n.risk n.event survival std.err lower 95% CI upper 95% CI
## 0 90 0 1.000 0.0000 1.000 1.000
## 1 81 9 0.900 0.0316 0.840 0.964
## 2 71 10 0.789 0.0430 0.709 0.878
## 3 60 11 0.667 0.0497 0.576 0.772
## 4 55 5 0.611 0.0514 0.518 0.721
## 5 44 6 0.543 0.0526 0.449 0.657## Warning: Vectorized input to `element_text()` is not officially supported.
## Results may be unexpected or may change in future versions of ggplot2.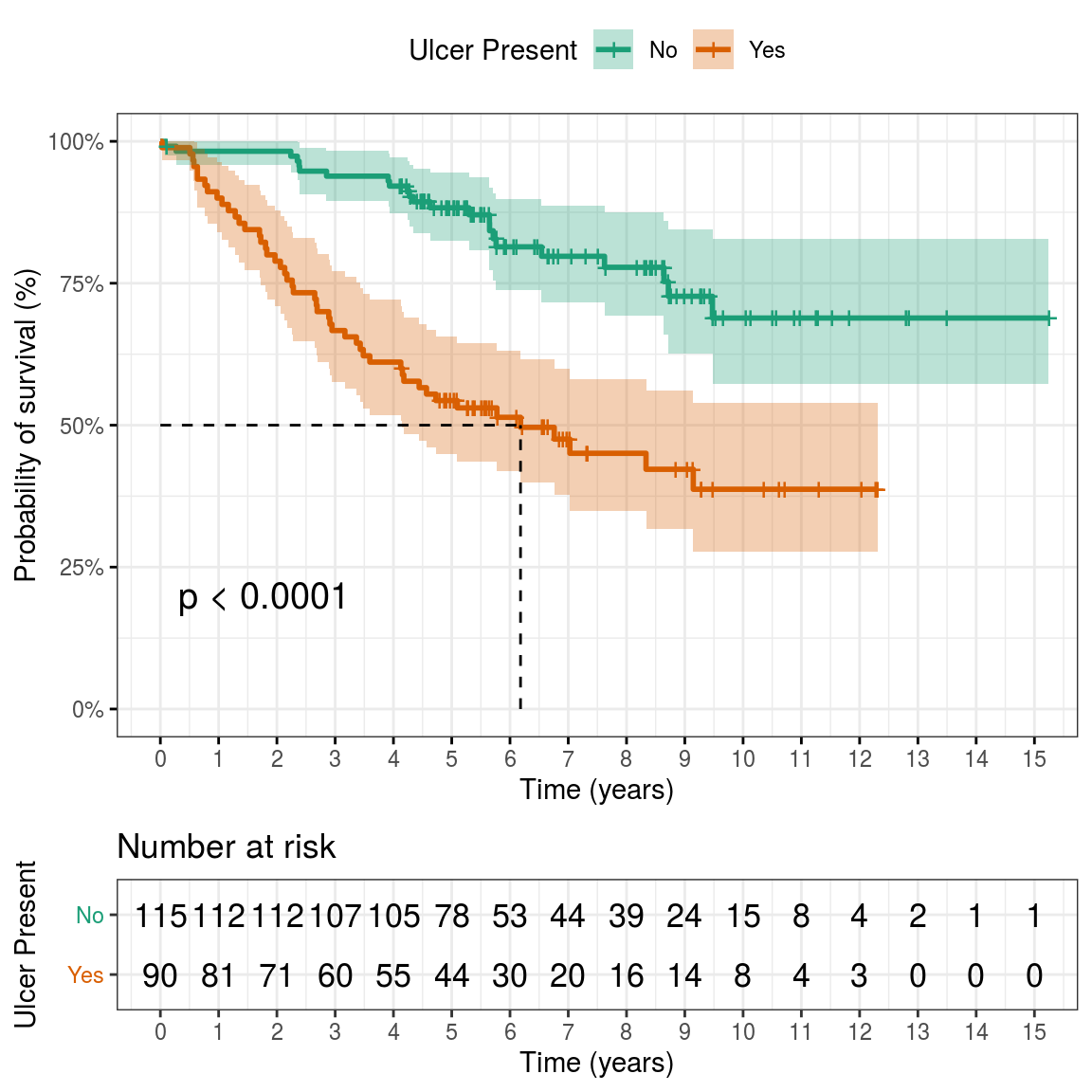
Solution to Exercise 10.12.2:
# Fit model
my_hazard = coxph(survival_object ~ sex + ulcer + age + thickness, data=melanoma)
summary(my_hazard)
# Melanoma thickness has a HR 1.11 (1.03 to 1.18).
# This is interpretted as a 11% increase in the
# risk of death at any time for each 1 mm increase in thickness.
# Check assumptions
ph = cox.zph(my_hazard)
ph
# GLOBAL shows no overall violation of assumptions.
# Plot Schoenfield residuals to evaluate PH
plot(ph, var=4)We also include a section on dealing with missing data in R. Something that we both feel strongly about and which is often poorly described and dealt with in academic publishing.