11.4 Look for patterns of missingness: missing_pattern()
Using finalfit, missing_pattern() wraps a function from the mice package, md.pattern().
This produces a table and a plot showing the pattern of missingness between variables.
explanatory <- c("age", "sex.factor",
"obstruct.factor",
"smoking_mcar", "smoking_mar")
dependent <- "mort_5yr"
colon_s %>%
missing_pattern(dependent, explanatory)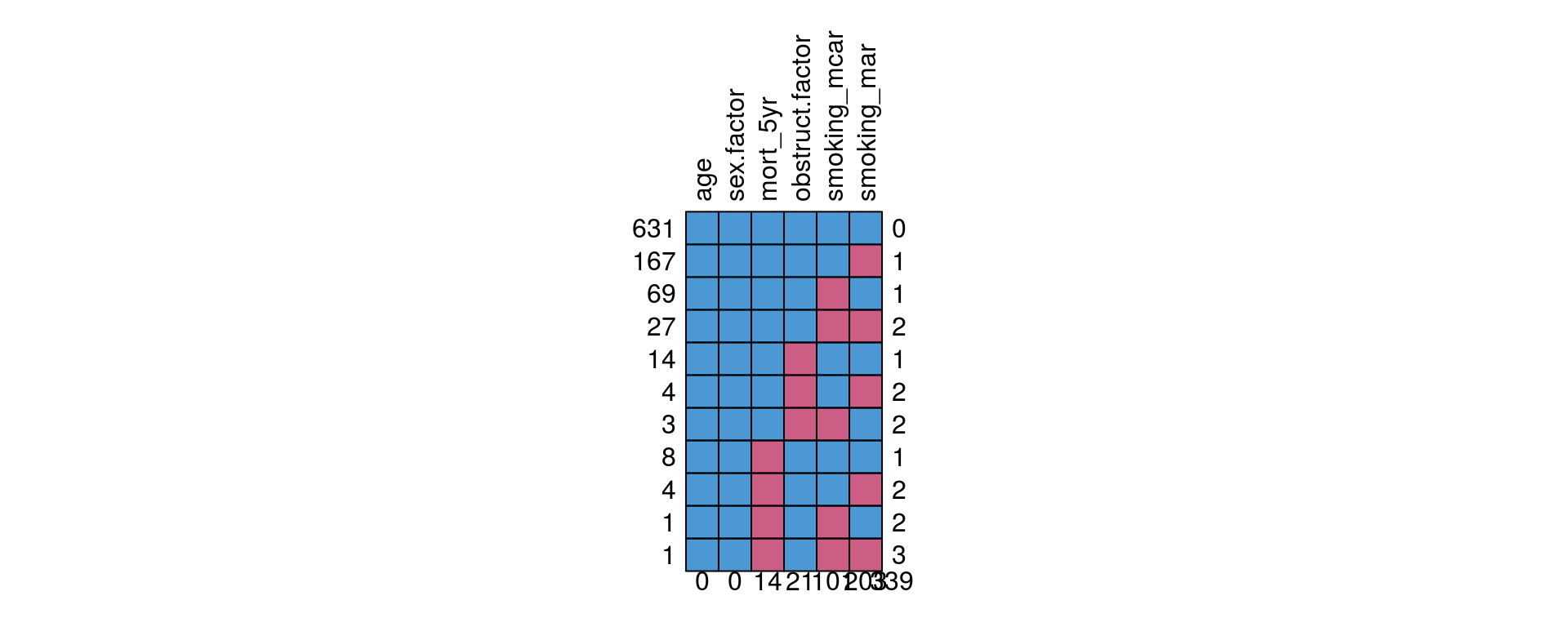
## age sex.factor mort_5yr obstruct.factor smoking_mcar smoking_mar
## 631 1 1 1 1 1 1 0
## 167 1 1 1 1 1 0 1
## 69 1 1 1 1 0 1 1
## 27 1 1 1 1 0 0 2
## 14 1 1 1 0 1 1 1
## 4 1 1 1 0 1 0 2
## 3 1 1 1 0 0 1 2
## 8 1 1 0 1 1 1 1
## 4 1 1 0 1 1 0 2
## 1 1 1 0 1 0 1 2
## 1 1 1 0 1 0 0 3
## 0 0 14 21 101 203 339This allows us to look for patterns of missingness between variables. There are 11 patterns in these data. The number and pattern of missingness help us to determine the likelihood of it being random rather than systematic.